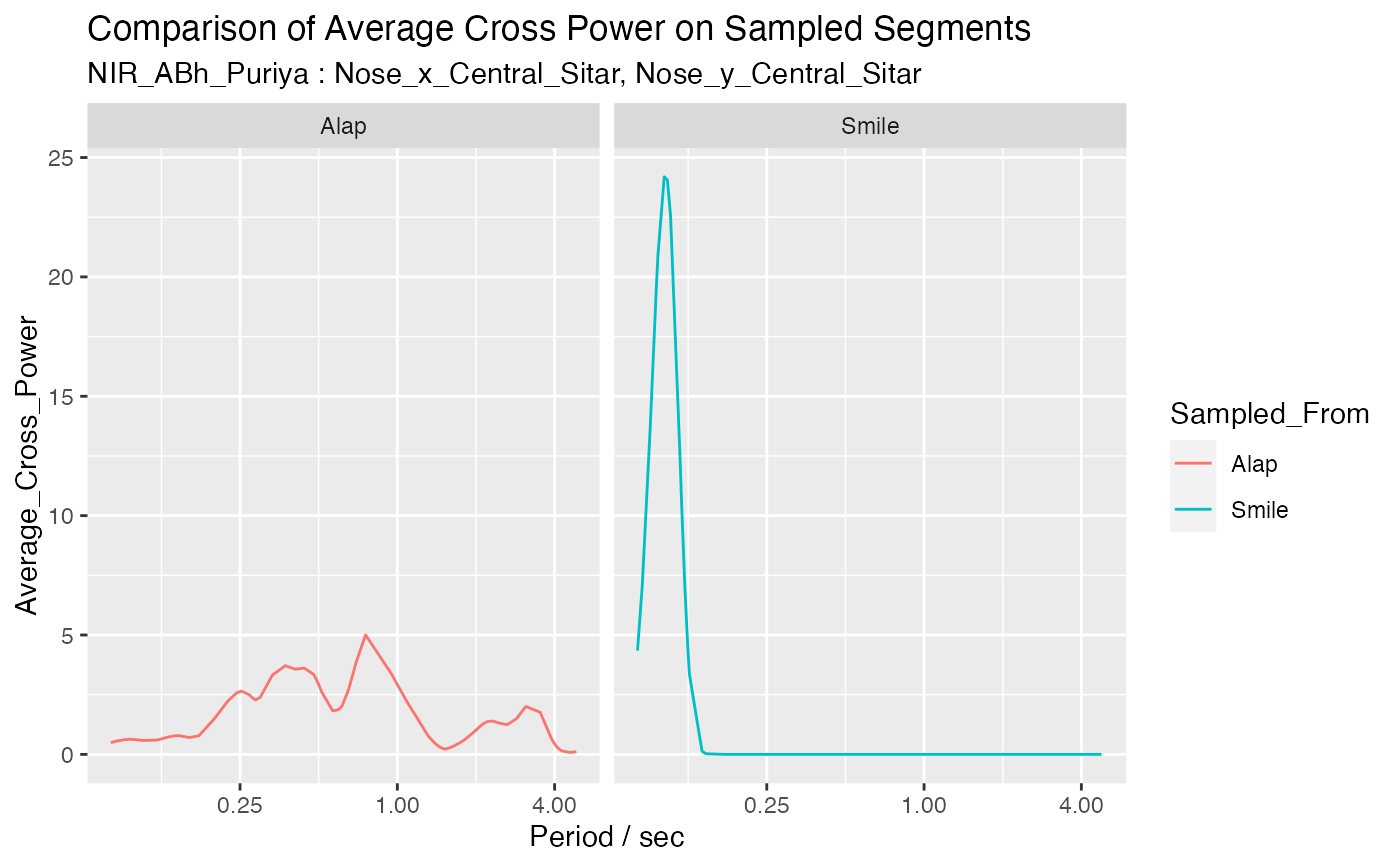
Compare the average cross power distribution of two SplicedViews using sampling on each segment
compare_avg_cross_power2.RdCompare the average cross power distribution of two SplicedViews using sampling on each segment
Arguments
- sv1
SplicedViewobject.- sv2
SplicedViewobject.- name1
name for first object.
- name2
name for second object.
- num_samples
number of samples to draw from segments.
- columns
column names in the data e.g. c('Nose_x', 'Nose_y').
- show_plot
show the plot?
See also
Other statistical and analysis functions:
apply_column_spliceview(),
apply_segment_spliceview(),
ave_cross_power_over_splices(),
ave_cross_power_spliceview(),
ave_power_over_splices(),
ave_power_spliceview(),
calculate_ave_cross_power1(),
calculate_ave_power1(),
compare_ave_cross_power1(),
compare_ave_power1(),
compare_avg_power2(),
difference_onsets(),
pull_segment_spliceview(),
sample_gap_splice(),
sample_offset_splice(),
summary_onsets(),
visualise_sample_splices()
Examples
r <- get_sample_recording()
d1 <- get_duration_annotation_data(r)
fv_list <- get_filtered_views(r, data_points = "Nose", n = 41, p = 3)
jv <- get_joined_view(fv_list)
# only one relevant section for sample data
splicing_smile_df <- splice_time(d1, tier ='INTERACTION',
comments = 'Mutual look and smile')
sv_duration_smile <- get_spliced_view(jv, splicing_df = splicing_smile_df)
splicing_alap_df <- splice_time(
d1, tier = 'FORM', comments = 'Alap'
)
sv_duration_alap <- get_spliced_view(jv, splicing_df = splicing_alap_df)
sample_list <- compare_avg_cross_power2(
sv_duration_smile, sv_duration_alap, 'Smile', 'Alap', num_samples = 100,
columns = c("Nose_x_Central_Sitar", "Nose_y_Central_Sitar"))
#> Warning: NaNs produced
#>
|
| | 0%
#> Warning: NaNs produced
#>
|
|======================================================================| 100%
#>
|
| | 0%
|
|======================================================================| 100%