Compare average cross power distribution using a splicing table
compare_ave_cross_power1.RdCompare average cross power distribution using a splicing table
Usage
compare_ave_cross_power1(
jv,
splicing_df,
splice_name,
num_segment_samples,
num_splice_samples,
columns,
sampling_type = "offset",
rejection_list = list(),
show_plot = TRUE
)Arguments
- jv
JoinedViewobject.- splicing_df
Spliceobject.- splice_name
Name to give randomly spliced segments.
- num_segment_samples
number of segments to randomly sample.
- num_splice_samples
number of randomly chosen splices.
- columns
name of data columns on which to calculate cross average power.
- sampling_type
either 'offset' or 'gap'.
- rejection_list
list of splice objects that random splices must not overlap.
- show_plot
show the plot? (Default is TRUE).
Value
list of two data frames: one containing average cross power on the first splice and the other containing the average cross power on randomly generated splices.
See also
Other statistical and analysis functions:
apply_column_spliceview(),
apply_segment_spliceview(),
ave_cross_power_over_splices(),
ave_cross_power_spliceview(),
ave_power_over_splices(),
ave_power_spliceview(),
calculate_ave_cross_power1(),
calculate_ave_power1(),
compare_ave_power1(),
compare_avg_cross_power2(),
compare_avg_power2(),
difference_onsets(),
pull_segment_spliceview(),
sample_gap_splice(),
sample_offset_splice(),
summary_onsets(),
visualise_sample_splices()
Examples
# \donttest{
r <- get_sample_recording()
fv_list <- get_filtered_views(r, data_points = 'Nose', n = 41, p = 3)
jv <- get_joined_view(fv_list)
splicing_df <- splice_time(list(a = c(0, 5), b = c(10, 15)))
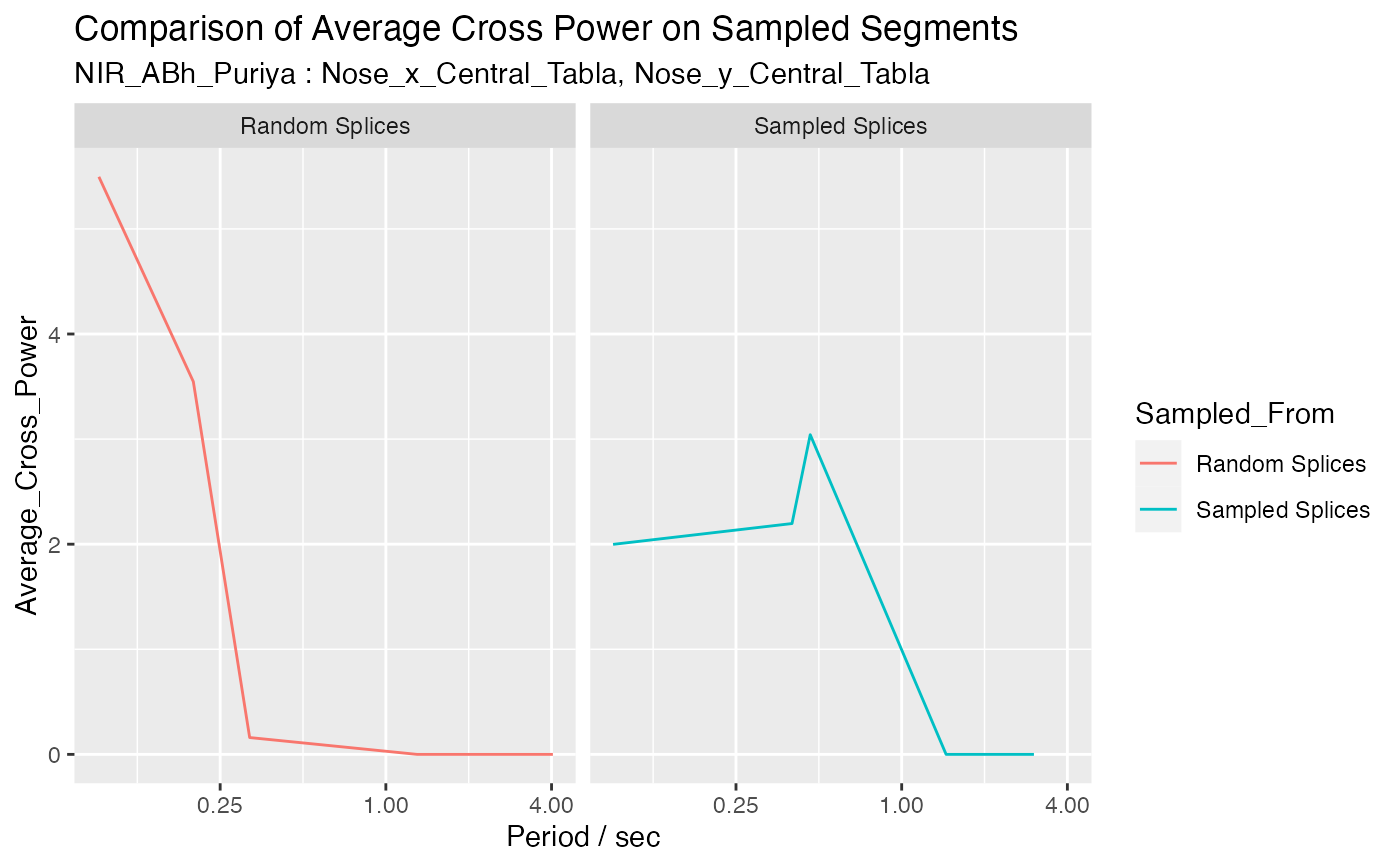
output_list <- compare_ave_cross_power1(jv, splicing_df, 'Random Splices', 5, 5,
c('Nose_x_Central_Tabla', 'Nose_y_Central_Tabla'))
#> Accepted splices: 4
#> Accepted splices: 7
#> Warning: NaNs produced
#>
|
| | 0%
#> Warning: NaNs produced
#>
|
|======================================================================| 100%
#> Warning: NaNs produced
#>
|
| | 0%
#> Warning: NaNs produced
#>
|
|======================================================================| 100%
#> Warning: NaNs produced
#>
|
| | 0%
#> Warning: NaNs produced
#>
|
|======================================================================| 100%
#> Warning: NaNs produced
#>
|
| | 0%
#> Warning: NaNs produced
#>
|
|======================================================================| 100%
#> Warning: NaNs produced
#>
|
| | 0%
#> Warning: NaNs produced
#>
|
|======================================================================| 100%
#> Warning: NaNs produced
#>
|
| | 0%
#> Warning: NaNs produced
#>
|
|======================================================================| 100%
#> Warning: NaNs produced
#>
|
| | 0%
#> Warning: NaNs produced
#>
|
|======================================================================| 100%
#> Warning: NaNs produced
#>
|
| | 0%
#> Warning: NaNs produced
#>
|
|======================================================================| 100%
#> Warning: NaNs produced
#>
|
| | 0%
#> Warning: NaNs produced
#>
|
|======================================================================| 100%
#> Warning: NaNs produced
#>
|
| | 0%
#> Warning: NaNs produced
#>
|
|======================================================================| 100%
#> Warning: NaNs produced
#>
|
| | 0%
#> Warning: NaNs produced
#>
|
|======================================================================| 100%
#> Warning: NaNs produced
#>
|
| | 0%
#> Warning: NaNs produced
#>
|
|======================================================================| 100%
 # }
# }