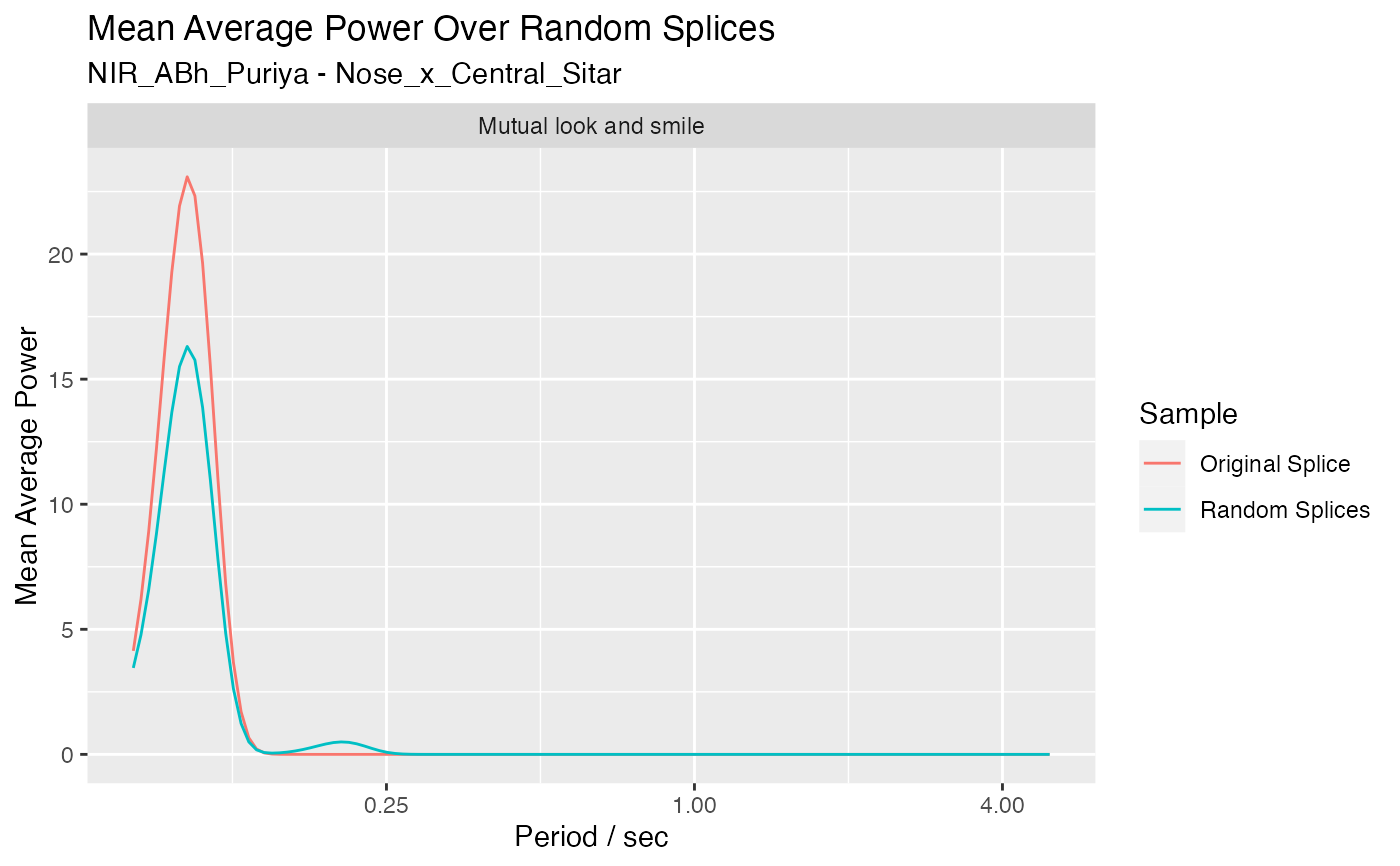
Calculate mean average power over splices using a splicing table
ave_power_over_splices.RdRandomly generates splices from a splicing table and calculates average power for each segment and splice. Calculates the mean average power over the random splices for each segment and period. Compares with the average power for the original splice.
Usage
ave_power_over_splices(
jv,
splicing_df,
num_splices,
column,
sampling_type = "offset",
rejection_list = list(),
include_original = TRUE,
show_plot = TRUE
)Arguments
- jv
JoinedViewobject.- splicing_df
Spliceobject.- num_splices
number of randomly chosen splices.
- column
name of data column on which to calculate average power.
- sampling_type
either 'offset' or 'gap'.
- rejection_list
list of splice objects that random splices must not overlap.
- include_original
include the original splice in output? (Default is TRUE).
- show_plot
show a plot? (Default is TRUE).
See also
Other statistical and analysis functions:
apply_column_spliceview(),
apply_segment_spliceview(),
ave_cross_power_over_splices(),
ave_cross_power_spliceview(),
ave_power_spliceview(),
calculate_ave_cross_power1(),
calculate_ave_power1(),
compare_ave_cross_power1(),
compare_ave_power1(),
compare_avg_cross_power2(),
compare_avg_power2(),
difference_onsets(),
pull_segment_spliceview(),
sample_gap_splice(),
sample_offset_splice(),
summary_onsets(),
visualise_sample_splices()
Examples
r <- get_sample_recording()
fv_list <- get_filtered_views(r, data_points = "Nose", n = 41, p = 3)
jv <- get_joined_view(fv_list)
d <- get_duration_annotation_data(r)
splicing_tabla_solo_df <- splice_time(d,
expr = "Tier == 'INTERACTION' & Comments == 'Mutual look and smile'")
# Only do the first splice for sample data
mean_ave_power_df <- ave_power_over_splices(jv, splicing_tabla_solo_df[1,], num_splices = 10,
column = 'Nose_x_Central_Sitar', show_plot = TRUE)
#> Accepted splices: 8
#> Accepted splices: 18
#> Starting wavelet transformation...
#> ... and simulations...
#>
|
| | 0%
|
|======================================================================| 100%
#> Class attributes are accessible through following names:
#> series loess.span dt dj Wave Phase Ampl Power Power.avg Power.pval Power.avg.pval Ridge Period Scale nc nr coi.1 coi.2 axis.1 axis.2 date.format date.tz
#> Starting wavelet transformation...
#> ... and simulations...
#>
|
| | 0%
|
|======================================================================| 100%
#> Class attributes are accessible through following names:
#> series loess.span dt dj Wave Phase Ampl Power Power.avg Power.pval Power.avg.pval Ridge Period Scale nc nr coi.1 coi.2 axis.1 axis.2 date.format date.tz
#> Starting wavelet transformation...
#> ... and simulations...
#>
|
| | 0%
|
|======================================================================| 100%
#> Class attributes are accessible through following names:
#> series loess.span dt dj Wave Phase Ampl Power Power.avg Power.pval Power.avg.pval Ridge Period Scale nc nr coi.1 coi.2 axis.1 axis.2 date.format date.tz
#> Starting wavelet transformation...
#> ... and simulations...
#>
|
| | 0%
|
|======================================================================| 100%
#> Class attributes are accessible through following names:
#> series loess.span dt dj Wave Phase Ampl Power Power.avg Power.pval Power.avg.pval Ridge Period Scale nc nr coi.1 coi.2 axis.1 axis.2 date.format date.tz
#> Starting wavelet transformation...
#> ... and simulations...
#>
|
| | 0%
|
|======================================================================| 100%
#> Class attributes are accessible through following names:
#> series loess.span dt dj Wave Phase Ampl Power Power.avg Power.pval Power.avg.pval Ridge Period Scale nc nr coi.1 coi.2 axis.1 axis.2 date.format date.tz
#> Starting wavelet transformation...
#> ... and simulations...
#>
|
| | 0%
|
|======================================================================| 100%
#> Class attributes are accessible through following names:
#> series loess.span dt dj Wave Phase Ampl Power Power.avg Power.pval Power.avg.pval Ridge Period Scale nc nr coi.1 coi.2 axis.1 axis.2 date.format date.tz
#> Starting wavelet transformation...
#> ... and simulations...
#>
|
| | 0%
|
|======================================================================| 100%
#> Class attributes are accessible through following names:
#> series loess.span dt dj Wave Phase Ampl Power Power.avg Power.pval Power.avg.pval Ridge Period Scale nc nr coi.1 coi.2 axis.1 axis.2 date.format date.tz
#> Starting wavelet transformation...
#> ... and simulations...
#>
|
| | 0%
|
|======================================================================| 100%
#> Class attributes are accessible through following names:
#> series loess.span dt dj Wave Phase Ampl Power Power.avg Power.pval Power.avg.pval Ridge Period Scale nc nr coi.1 coi.2 axis.1 axis.2 date.format date.tz
#> Starting wavelet transformation...
#> ... and simulations...
#>
|
| | 0%
|
|======================================================================| 100%
#> Class attributes are accessible through following names:
#> series loess.span dt dj Wave Phase Ampl Power Power.avg Power.pval Power.avg.pval Ridge Period Scale nc nr coi.1 coi.2 axis.1 axis.2 date.format date.tz
#> Starting wavelet transformation...
#> ... and simulations...
#>
|
| | 0%
|
|======================================================================| 100%
#> Class attributes are accessible through following names:
#> series loess.span dt dj Wave Phase Ampl Power Power.avg Power.pval Power.avg.pval Ridge Period Scale nc nr coi.1 coi.2 axis.1 axis.2 date.format date.tz
#> Starting wavelet transformation...
#> ... and simulations...
#>
|
| | 0%
|
|======================================================================| 100%
#> Class attributes are accessible through following names:
#> series loess.span dt dj Wave Phase Ampl Power Power.avg Power.pval Power.avg.pval Ridge Period Scale nc nr coi.1 coi.2 axis.1 axis.2 date.format date.tz