# read study encodings
study_encodings <- read.delim(
'bib/extractions.bib',
sep = '@',
header = F,
na.strings = c("NA", "")
)
# the citekeys appear in the second column
citekey <- study_encodings[,2]
citekey <- citekey[!is.na(citekey)]
# remove all but FEATURE_CATEGORIES ASCII field
feature_class <- study_encodings[,1]
feature_class <- feature_class[
stringr::str_detect(
feature_class,
"FEATURE_CATEGORIES"
)
]
feature_class <- na.omit(feature_class)Feature Representation
This notebook prepares a data frame tabulating the classes of features for each study included in the meta analysis. Feature categories come from Panda et al. (2020).
Procedure
To tabulate feature classes, we perform the following steps: 1. Read in .bib file and select only relevant fields. 2. Create and clean a data frame containing citation keys and feature categories 3. Account for discrepancies in encoding of features 4. Populate columns for each category tracking whether it was examined in study
1. Reading in bib file
First we read in the bibtex file and do some preprocessing to identify NA rows, along with the ASCII fields of interest.
2. Prepare dataframe
Next we combine the feature categories and citation keys into a data frame. We clean columns to account for excess characters from the bib file.
df_features <- data.frame(cbind(feature_class, citekey))
# remove additional characters from .bib file, add formatting
df_features[,1] <- stringr::str_extract(
df_features[,1],
"(?<=\\{).+?(?=\\})"
) |> tolower()
df_features[,1] <- stringr::str_remove_all(
df_features[,1],
",|;"
)
df_features[,2] <- stringr::str_remove_all(
df_features[,2],
"Article\\{|,"
)3. Account for discrepancies
Now we remove studies with no feature categories included or those lacking sufficient detail.
# remove unencoded studies
df_features <- subset(
df_features,
!feature_class %in% c("uncertain", "not specified")
)
df_features <- dplyr::filter(
df_features,
feature_class != " "
)4. Standardize feature categories
We’ll format names so they’re consistent when we tabulate feature categories in the next step. In some cases categories might be encoded as “melodic” and in others “melody”, but these refer to the same class, so we need to standardize them.
# simplify griffiths et al. (2021)
df_features[
df_features$citekey == "griffiths2021am",
]$feature_class <- "dynamic timbre harmony rhythm"
# standardize names
df_features[,1] <- stringr::str_replace_all(df_features[,1], "pitch", "melodic")
df_features[,1] <- stringr::str_replace_all(df_features[,1], "melodic", "melody")
df_features[,1] <- stringr::str_replace_all(df_features[,1], "harmonic", "harmony")
df_features[,1] <- stringr::str_replace_all(df_features[,1], "dynamics", "dynamic")5. Populate columns
Now we’ll create a column for each category and tabulate whether it was included for each study.
First we identify all unique category names:
# split encoding into a vector of values delimited by a space
category_representation <- strsplit(df_features[,1], " ")
# identify unique values
categories <- unique(
unlist(
category_representation
)
)Next we populate empty columns for each category name and combine these with our existing dataframe.
# populate a matrix of category names with zeroes
category_matrix <- matrix(
ncol = length(categories),)
category_matrix[1:length(categories)] <- 0
colnames(category_matrix) <- categories
df_features <- data.frame(
df_features,
category_matrix
)Now we’ll loop through the names of each feature category and identify whether it’s present in the “feature_class” column using the stringr package. This will add a “1” to the relevant column if the feature class is represented in a study.
for(this_category in categories) {
df_features[,this_category] <- stringr::str_count(
df_features$feature_class,
this_category
)
}After verifying this works, we no longer need the feature_class column, so we can remove it.
df_features$feature_class <- NULLWe can also remove rows corresponding to studies not included in the final selection of the metaanalysis.
# read in citekeys for included studies
included_citekeys <- c(
unique(read.csv("../analysis/R_summary.csv")$citekey),
unique(read.csv("../analysis/C_summary.csv")$citekey)
)
# subset only included studies
df_features <- subset(
df_features,
citekey %in% included_citekeys
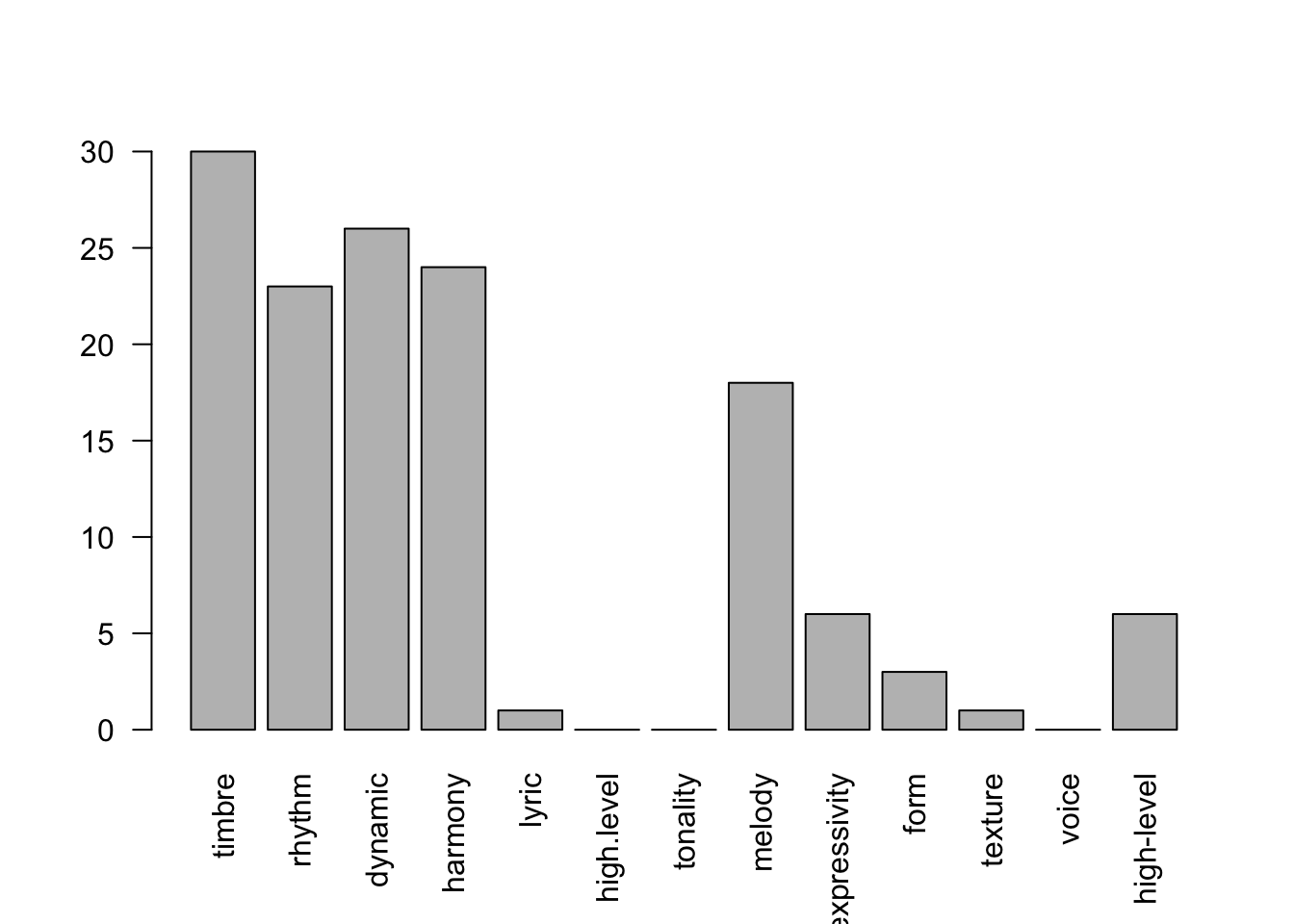
)Now we can see the common feature classes included in MER studies
barplot(
sapply(
df_features[,-1],
sum
),
las = 2
)
References
Panda, R., Malheiro, R., & Paiva, R. P. (2020). Audio features for music emotion recognition: a survey. IEEE Transactions on Affective Computing, 14(1), 68-88.